The approachModuleAlign uses a novel scoring scheme that integrates sequence information and both local and global network topology. It computes a homology score between proteins based on a hierarchical clustering of the input networks. (a) shows the hierarchical structure of a network. Leaves represent the proteins and each cluster joins sets of proteins with similar interactions. Starting from an optimal bipartite matching, ModuleAlign applies an iterative approach to find an alignment that scores high in our model while trying to preserve interactions. (b) shows the iterative refinement strategy of the alignment. Blue dotted lines represent the current alignment. First, a pair of proteins with largest score, shown in red, is selected. One of the neighbors of ub, shown in blue, is selected and its mapping is removed from the alignment. Next the alignment scores between u and all neighbors of vb are upweighted (green arrows) to promote the preserveation of interactions. While fixing the edge between the highest scoring pair of proteins (red), ModuleAlign runs one primal-dual iteration of the Hungarian method to update the alignment (see paper for details). | 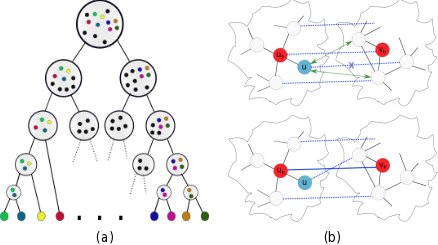 |